Biosensors
- Post by: Leo
- January 6, 2021
- Comments off
Biosensors: Bacteria detection, antibacterial resistance, chemicals, and toxins
Works have been accomplished…
-
Design and fabricate a biosensor to detect and quantify natural phenolic antioxidant with anticancer medicinal properties
-
Design and fabricate a label-free DNA-based biosensor to evaluate the efficacy of anticancer drugs (e.g., epirubicin) and their side effects
-
Pathogenic bacteria detection using droplet microfluidics vs. PCR
-
Design and fabricate a microfluidic platform for antibiotic susceptibility testing
-
In situ and ex situ sweat analysis: A versatile, cost-effective, and flexible wearable biosensor
-
Electrolytes (e.g., sodium and potassium), metabolites (e.g., lactate and pyruvate), nitrogenous substances (e.g., amino acids and proteins), nutrients (e.g., glucose and vitamins), and various disease biomarkers
-
A novel sensor for the detection of TiO2 nanoparticles from industrial, agricultural, pharmaceutical and medical fields
-
Design and fabricate implantable sensors that detect biomarkers for early disease diagnostics
-
Design and fabricate microfluidic platform to screen DNA library to gain insights on a linkage between phenotype and genotype
-
Develop a label-free single-cell protein quantification method using a drop-based mix-and-read system
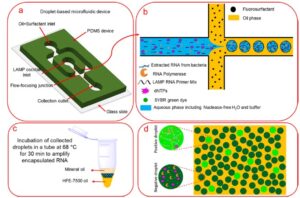
Schematic representations of sample encapsulation, incubation, and screening. (a) The schematic of the microfluidic flow-focusing device for encapsulation of the LAMP reaction cocktail into water-in-oil droplets. (b) Schematic view of the flow-focusing cross-junction. The water-in-oil droplets are collected into a PCR tube. (c) Incubation of collected droplets in a thermocycler at 68 °C for 30 min. (d) Fluorescent imaging and “Yes/No” quantification of the incubated positive droplets.
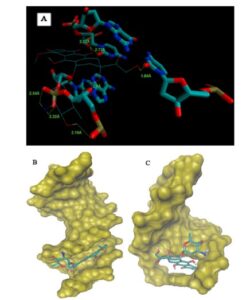
(A) Geometrical disposition of epirubicine in DNA intercalation distances in A. (B) Epirubicin DNA minor groove interaction. (C) Epirubicin DNA major groove interaction.
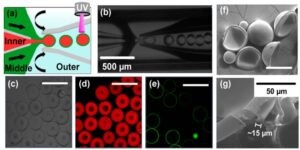
Microcapsules fabrication. (a) Illustration of the capillary microfluidic device for fabrication of microcapsules. The inner (red, water phase containing nanosensors) and middle (green, oil phase containing PEGDA monomer) fluids were focused by the outer (blue, water phase) fluid to generate double emulsion droplets. (b) Optical microscope image showing double emulsion droplet formation. (c) Optical image of the as-fabricated microcapsules. (d) QDs (red) were encapsulated in the capsules. (e) Capsule shells were stained with green fluorescent dye. (f) SEM images of dried microcapsules and (g) the cross-section of the capsule shell was shown. Scale bar: 500 μm in panels c−e and 200 μm in panel f.
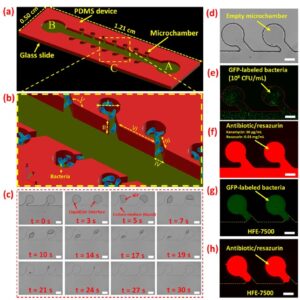
Schematic and experimental representations of N-3M platform for AST assay. (a) The schematic structure of the N-3M device. A and B are inlet and outlet, respectively. (b) The schematic AST testing zone is magnified. The I, II, III, IV, V, and VI set is 200 μm, 40 μm, 100 μm, 135°, and 400 μm, respectively. (c) Sequence of bright-field images showing the bacterial suspension loading process. (d) Two consecutive empty microchambers. Microchambers are loaded with (e) bacterial suspension and then (f) antibiotic/resazurin mixture solutions. Flushing the excess bacteria suspension and antibiotic/resazurin from the main channel and isolating microchambers with HFE-7500 in (g) green and (h) red fluorescent modes. Scale bars are 100 μm.
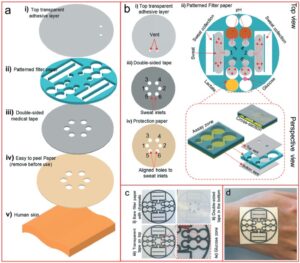
Schematic design of the wearable device for sweat collection and bio-sensing. (a) Schematic illustration of the wearable POC device layers, including the (i) top transparent adhesive layer, (ii) the pattern supported on filter paper, (iii) the double-sided medical tape, and (iv) a protective piece paper, which when fully assembled can serve as a patch that can be mounted on (v) human skin. (b) The device functionality in more detail, including: (i) two vents on the transparent adhesive layer for enzymatic O2 delivery for the glucose and lactate oxidation reactions; (ii) the full design for collecting sweat and monitoring pH, glucose, and lactate, all supported on filter paper (the magnified inset illustrates the glucose detection zone and cross-sectional view of the design); (iii) the double-sided tape; and (iv) the protective paper. (c) Images of the device (diameter: 3.9 cm) before and after the addition of the transparent and medical tape layers, along with a magnified image of the glucose detection zone. (d) Image of the wearable device attached on a human hand.
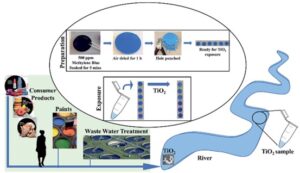
Schematic illustration of the preparation of the paper-based functional device for TiO2 sensing
Evaluate the use of a rapid, low-cost, portable strategy for monitoring changes in color of vapochromic materials during exposure to chemical vapors.
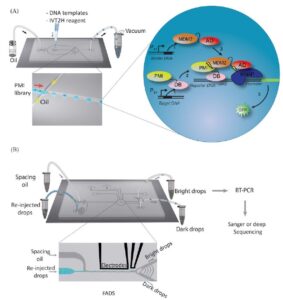
A schematic workflow for screening high-affinity binders using drop-based in vitro two hybrid method (drop-IVT2H). (A) Single DNA templates and IVT2H reagents were encapsulated in drops on a microfluidic chip by applying vacuum to generate monodisperse drops. The binder DNA template was distributed as single copy in drops based on Poisson statistics. Each drop contained multiple copies of the reporter and target DNA templates. Drops were then collected for off-chip incubation. Inset, the IVT2H reaction during off-chip incubation. PMI-DB and AD-MDM2 were expressed from the binder and target templates, respectively. PMI-DB bound the upstream activation sequence (UAS) on the reporter DNA. The interaction of PMI and MDM2 recruited AD to the promoter-bound RNA polymerase (RNAP) and activated the expression of the reporter gene, producing the fluorescent GFP. (B) After incubation, drops were re-injected into a fluorescence-activated drop sorting device (FADS). Co-flowing spacing oil ensured equal separation of drops. Both bright and dark drops were isolated and collected in separate tubes for RT-PCR followed by Sanger or deep sequencing.
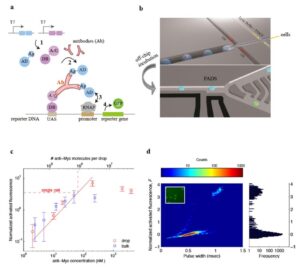
(a) Scheme of PAIGE for detecting an antigen (Ag)-specific antibody (Ab). Two fusion proteins, AD-Ag and A/G-DB, are constitutively expressed under T7 promoter (T7) from input DNA templates by T7 RNA polymerase (step 1). The antibody binds to both Ag and A/G, forming a ternary complex that binds to the upstream-activation sequence (UAS) on the reporter DNA and recruits AD near the promoter-bound RNA polymerase (RNAP) (step 2). AD activates RNAP (step 3) to express the reporter gene, which produces GFP (step 4). (b) The workflow of drop-based PAIGE. Single cells (megenta) are encapsulated with PAIGE and a lysis buffer in drops (blue). After off-chip incubation, drops are re-injected into a fluorescence-activated drop-sorting (FADS) device to measure their fluorescence and sort them based on a fluorescence threshold. (c) Titration curves of pure anti-myc in microwell (blue square) and drop-based (red circle) PAIGE. The dashed line shows the average number of anti-Myc molecules in a single cell. The fluorescence of anti-Myc in microwells or drops is normalized by that of microwells or drops without anti-Myc. The normalized activated fluorescence, F, is obtained by subtracting one. (d) Heat map showing the distribution of drops in terms of their normalized activated fluorescence, F, and width recorded as the PMT’s pulse duration (left). The corresponding fluorescence histogram is shown on the right. The inset fluorescence microscopy image shows that after incubation a small fraction of the drops is bright.
Peer-reviewed Publications
-
Nanoliter-Sized Microchamber/Microarray Microfluidic Platform for Antibiotic Susceptibility Testing (https://doi.org/10.1021/acs.analchem.8b03817)
-
Pathogenic Bacteria Detection Using RNA-Based Loop-Mediated Isothermal-Amplification-Assisted Nucleic Acid Amplification via Droplet Microfluidics (https://doi.org/10.1021/acssensors.8b01206)
-
A new epirubicin biosensor based on amplifying DNA interactions with polypyrrole and nitrogen-doped reduced graphene: experimental and docking theoretical investigations (https://doi.org/10.1016/j.snb.2018.12.164)
-
A versatile, cost-effective, and flexible wearable biosensor for in situ and ex situ sweat analysis, and personalized nutrition assessment (10.1039/c9lc00734b)
-
Determination of ferulic acid in the presence of butylated hydroxytoluene as two phenolic antioxidants using a highly conductive food nanostructure electrochemical sensor (https://doi.org/10.1007/s11696-019-00793-y)
-
A novel paper based colorimetric assay for the detection of TiO2 nanoparticles (10.1039/C7AY02700A)
-
Microfluidic fabrication of colloidal nanomaterials-encapsulated microcapsules for biomolecular sensing (https://doi.org/10.1021/acs.nanolett.7b00026)
-
Label-free single-cell protein quantification using a drop-based mix-and-read system (https://doi.org/10.1038/srep12756)
-
A mix-and-read drop-based in vitrotwo-hybrid method for screening high-affinity peptide binders (https://doi.org/10.1038/srep22575)
-
A digital imaging method for evaluating the kinetics of vapochromic response (https://doi.org/10.1016/j.talanta.2019.120520)
Categories: