Gut on a chip
- Post by: Leo
- January 6, 2021
- Comments off
Gut on a Chip
Works have been accomplished…
-
A microfluidic-based model incorporating bacteria embedded hydrogel microfibers for the coculture of human enteric bacteria
-
Study the inflammatory bowel diseases such as Crohn’s disease
-
Metabolite analysis of the medium
-
Decipher the cross-talk within the polymicrobial intestinal luminal environment, and its impact on the intestinal epithelium
-
3D microenvironment for bacterial cell growth and proliferation: Mimic cell natural growth
-
Microfluidic-based cell encapsulation
-
Study cellular dynamics and interactions
-
Study the interaction between crypt cells and Peyer’s patch immune cells
-
Study intercellular chemical interactions
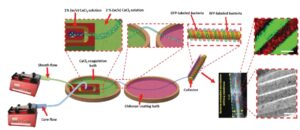
A schematic design of the fiber generating microfluidic flow-focusing device, coagulation and coating baths, and the winding machine. Inset images include the bacterial-embedded fibrous membrane composed of 50 winds of fiber, the bright field and fluorescent images of bacterial embedded fibers (scale bars are 100 µm).
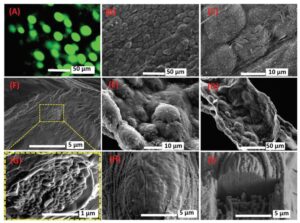
A) Fluorescent image of cultured GFP-labeled E. coli inside alginate/CMC/mucin microfibers after 60 h culture in DMEM medium at room temperature (22 ± 2 °C). SEM images of the vacuum dried bacteria-embedded microfibers at different magnifications, including B) 540× and C) 3000×, and SEM images displaying the morphology of the longitudinal cut of the freeze-dried bacteria-embedded microfibers at two magnifications, including D) 600× and E) 2200×. Cryo-SEM images of the cross-section of F) the bacterial cell-embedded microfiber and G) a bacterial nodule. Cryo-SEM images of H) the surface of a bacterial nodule and I) a FIB sectioned nodule.
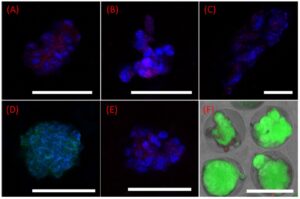
Confocal images of intestinal organoids within microgels cocultured with Peyer’s patch cells in the 3D-printed insert. Bmi-1 (red) and DAPI (blue) staining of organoids cultured in crypt niche microgels (alginate/gelatin 1:1, containing fibronectin, collagen IV, and laminin proteins) on (A,E) day 7, (B) day 14, and (C) day 21. (D) Villin (green) and DAPI (blue) staining of the organoids on day 7. (F) Live (green) and dead (red) staining of crypt cells within microgels cocultured with Peyer’s patch cells in the 3D-printed insert on day 21. The scale bar is 70 μm.
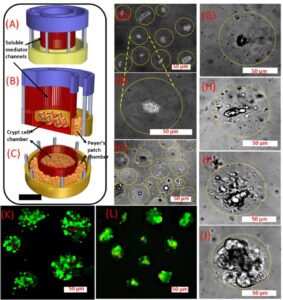
(A−C) 3D-printed insert for coculture of the crypt and Peyer’s patch cell-embedded microgels. (D,E) Encapsulated crypt cells inside alginate/gelatin (1:1) microgels. (F) Peyer’s patch cell-embedded alginate/gelatin microgel (1:1). (G−J) Crypt cell growth inside the microgels. Isolated crypt cells from a mouse intestine, cultured in the 3D-printed insert on (G) day 1, (H) day 3, (I) day 7, and (J) day 14 along with Peyer’s patch cells. The borders of the microgels have been encircled inside dotted lines. Live (green) and dead (red) staining of (K) Peyer’s patch cells after 7 days cocultured with (L) crypt cells.
Peer-reviewed Publications
-
Microfluidic-based cell-embedded microgels using nonfluorinated oil as a model for the gastrointestinal niche (https://doi.org/10.1021/acsami.7b16916)
-
A Microfluidic‐Based Model for Spatially Constrained Culture of Intestinal Microbiota (https://doi.org/10.1002/adfm.201805568)
Categories: